Build, Run, Visualize#
The Python examples do not need to be built. First, install Python from python.org, Anaconda, or a system package manager like Ubuntu Linux apt or macOS Homebrew.
Next, install the itk package:
python -m pip install --upgrade pip
python -m pip install itk
Download an example by clicking the Download button.#
Download the <ExampleName>.zip file from the link on the top of an example’s webpage. Unpack the example. For example,
unzip ExampleName.zip
Finally, run the example:
python ExampleName/Code.py
Build individual examples#
Download the <ExampleName>.zip file from the link on the top of the example’s webpage. Unpack the example:
unzip ExampleName.zip
Set convenience variables:
ITK_SOURCE=ExampleName
ITK_BUILD=ExampleName/build
Move to the build directory:
cd ${ITK_BUILD}
Run CMake (minimum version 3.10.2) to configure the project.
If ITK Version 5.0.0 or above not installed but compiled on your system, you will need to specify the path to your ITK build:
cmake -DITK_DIR=/home/luis/itk_build -S ${ITK_SOURCE} -B ${ITK_BUILD}
Build the project, and run the test:
cmake --build .
ctest -V
Build all examples and the documentation#
Set convenience variables:
ITK_SOURCE=ITKEx
ITK_BUILD=ITKEx-build
Clone the repository:
git clone --recursive https://github.com/InsightSoftwareConsortium/ITKSphinxExamples.git ${ITK_SOURCE}
Make a build directory:
mkdir -p ${ITK_BUILD}
cd ${ITK_BUILD}
Run CMake (minimum version 3.10.2) to configure the project.
If ITK is not installed nor compiled, you can then make use of the superbuild functionality:
cmake -DBUILD_DOCUMENTATION:BOOL=ON -S ${ITK_SOURCE}/Superbuild/ -B ${ITK_BUILD}
If ITK (Version 5.0.0 or above) is installed:
cmake -DBUILD_DOCUMENTATION:BOOL=ON -S ${ITK_SOURCE} -B ${ITK_BUILD}
If ITK (Version 5.0.0 or above) is not installed but compiled on your system, you will need to specify the path to your ITK build:
ITK_COMPILED_DIR=/usr/opt/itk_build cmake -DITK_DIR=${ITK_COMPILED_DIR} -DBUILD_DOCUMENTATION:BOOL=ON -S ${ITK_SOURCE_DIR} -B ${ITK_BUILD_DIR}
The superbuild will download and build all required dependencies. If you are building the documentation and not using superbuild functionality, then you must have all the required dependencies installed, which are listed in the README.rst file located at the root of the source tree. If you just want to build the examples and not their documentation, set BUILD_DOCUMENTATION to OFF in your CMake configuration.
Build the project (this will generate the documentation and all examples):
cmake --build .
Run the tests with a superbuild:
cd ${ITK_BUILD_DIR}/ITKEX-build
ctest -V
Run the tests without a superbuild:
cd ${ITK_BUILD_DIR}
ctest -V
Run an example#
After building the examples, you can run an example by using cd to move to the example’s directory. Then, directly run the executable.
Alternatively, the ctest command line program that comes with CMake can be used to drive the examples as unit test. Running:
ctest
in the binary tree will run all unit tests found in the current directory and below.
ctest -R Binary
will run all tests whose name matches the regular expression Binary.
ctest -V
will run ctest in verbose mode, which prints out the command executed and all of the resulting text output.
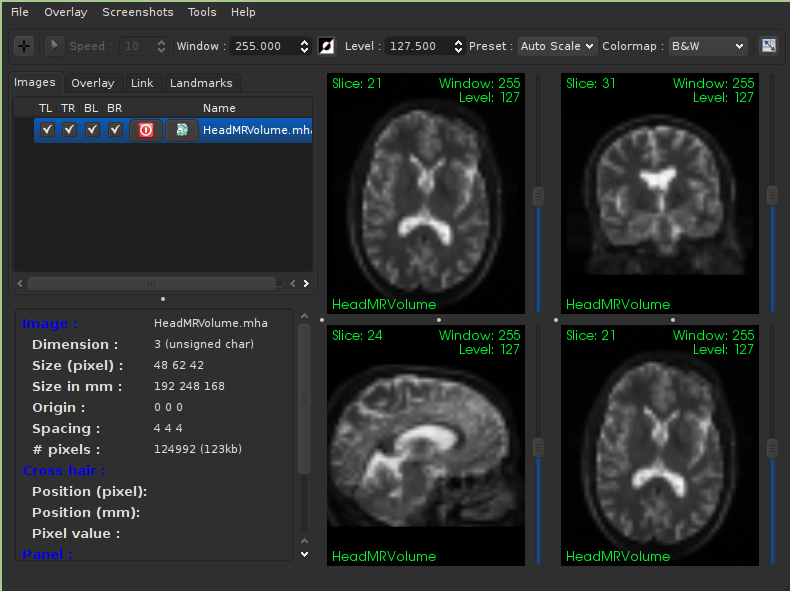
Visualize the results#
ITK is a library limited in scope to image analysis, and it purposely does not attempt to perform image visualization. Visualizing the results of analysis is possible with a number of third-party applications. Note that these packages are specially suited for medical images, which often have anisotropic spacing and can span three or more dimensions. All applications listed are open source and cross-platform.
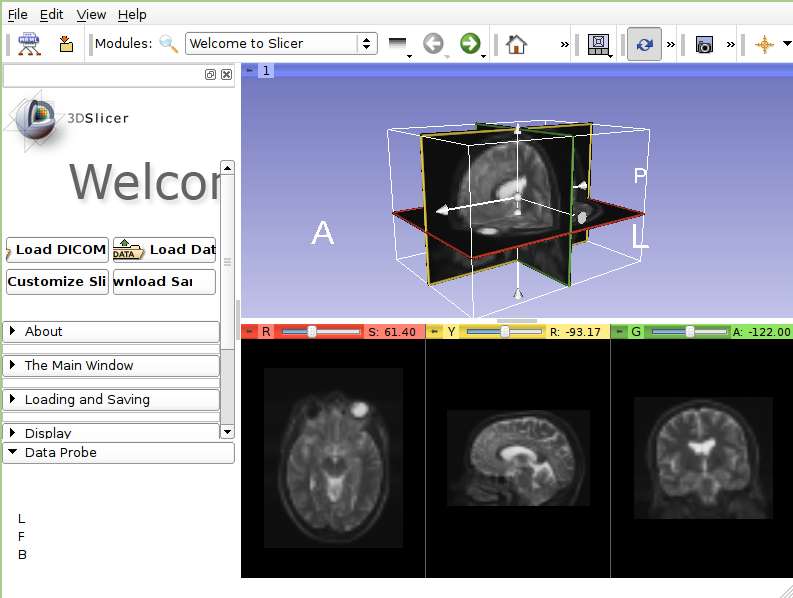
- 3DSlicer
3DSlicer is an open-source software platform for the analysis and visualization of medical images and for research in image guided therapy. The platform provides functionality for segmentation, registration and three-dimensional visualization of multi-modal image data, as well as advanced image analysis algorithms for diffusion tensor imaging, functional magnetic resonance imaging and image-guided therapy. Standard image file formats are supported, and the application integrates interface capabilities to biomedical research software and image informatics frameworks.
- ImageViewer
An FLTK-based ImageViewer was extracted from the ITKApps repository. This simple yet effective slice-based viewer works on 2D and 3D images and supports probing of data values.
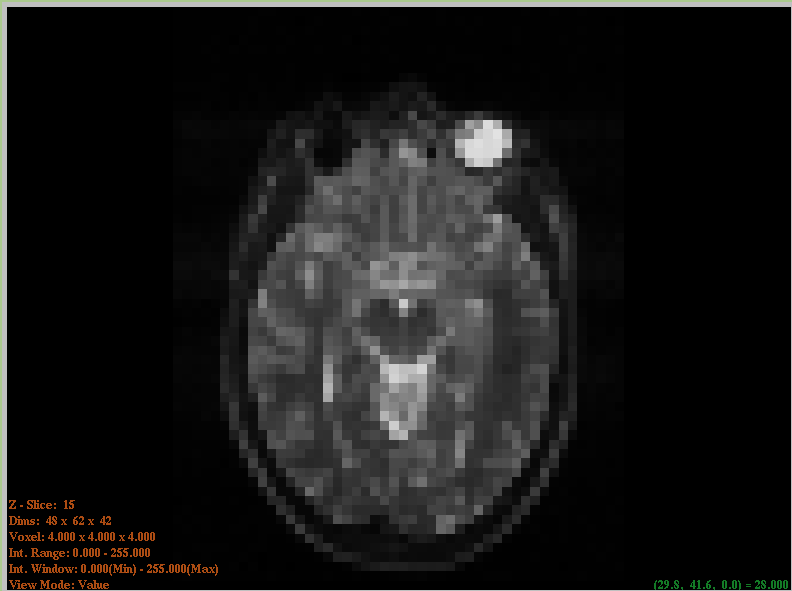
ImageViewer#
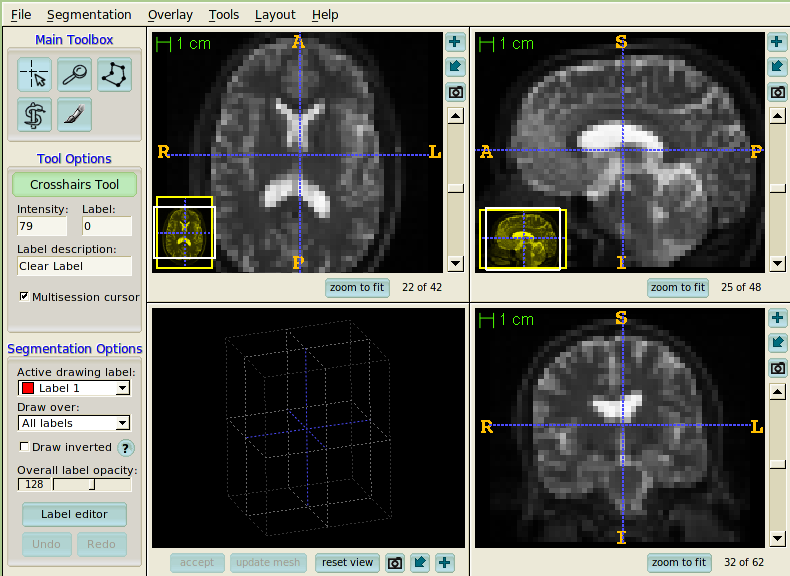
- ITK-SNAP
ITK-SNAP is segmentation application, but it is also a nice general resource for visualization of the results of analysis.
- MITK
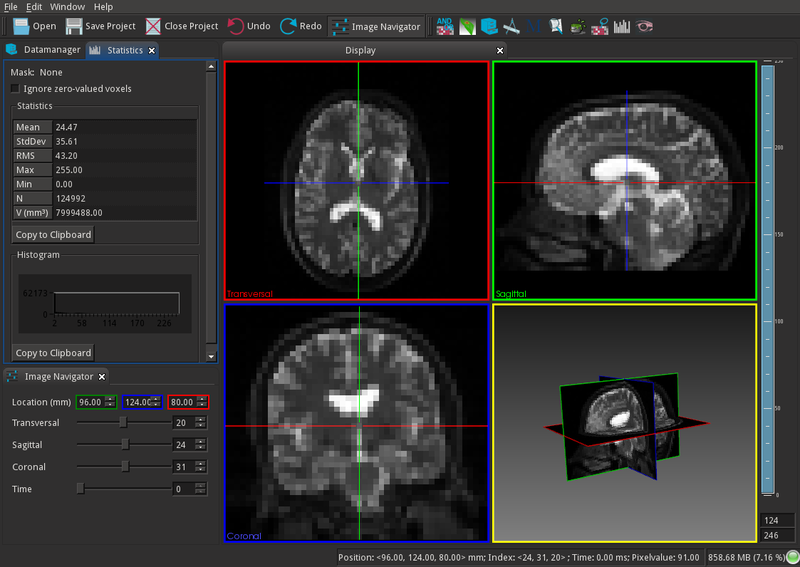
MITK is a free open-source software system for development of interactive medical image processing software.
- Paraview
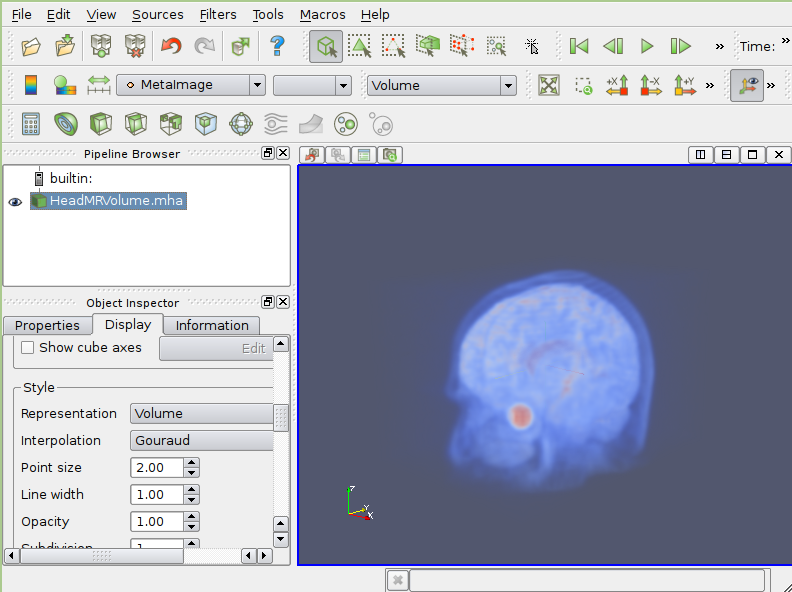
Paraview is a full-featured scientific visualizion GUI written with Qt/VTK. It has extensive parallel processing capabilities.
- VV
VV is an image viewer designed for fast and simple visualization of spatio-temporal images: 2D, 2D+t, 3D and 3D+t (or 4D) images.